Overview
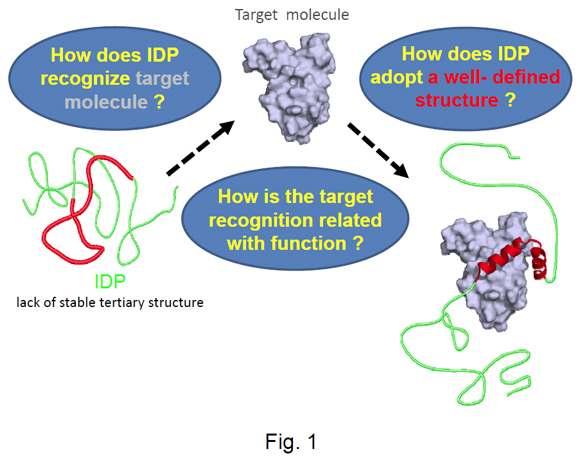
Separated from their binding partners, intrinsically disordered proteins (IDPs) show regions of no stable structure. These regions adopt well-defined structures only upon binding to their targets (Fig. 1). This behavior differs significantly from well-characterised types of interaction between proteins such as induced-fit, in which stable structures bend slightly to accommodate each other. Our research project is expected to establish a new paradigm for protein structure and function. IDPs are mostly found in eukaryotes, and generally interact with several target molecules. They are often important protein hubs within intracellular protein interaction networks. Understanding IDPs is therefore of great importance to investigations of cellular functions, and for developing new drugs intended to disrupt signalling networks in eukaryotic cells.
We have divided the research into the following three research projects (Fig. 2); 1) development of new methods to determine IDP structures showing very large fluctuations, 2) functional analysis of IDPs by means of molecular biology, biochemistry and molecular genetics, and 3) prediction of structure and function of IDP by means of computer simulation and bioinformatics. Together the three projects are aimed at understanding the target recognition mechanism of IDPs (Fig. 2).

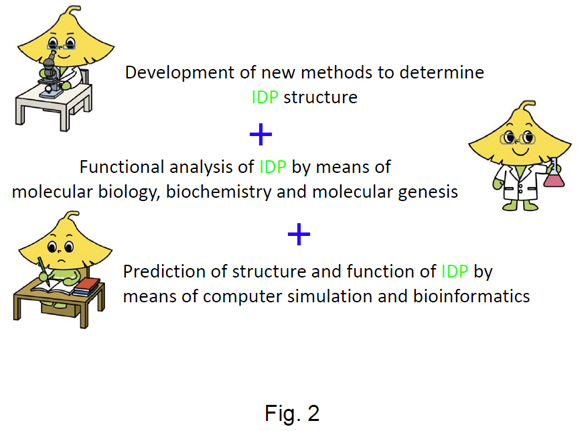
We have divided the research into the following three research projects (Fig. 2); 1) development of new methods to determine IDP structures showing very large fluctuations, 2) functional analysis of IDPs by means of molecular biology, biochemistry and molecular genetics, and 3) prediction of structure and function of IDP by means of computer simulation and bioinformatics. Together the three projects are aimed at understanding the target recognition mechanism of IDPs (Fig. 2).

