The mucosal epithelium that lines the inner surfaces of the body, especially within the intestine, is exposed to various macromolecules and microorganisms whose efficient uptake is crucial to maintaining normal immune regulation. Epithelial cells that cover the gut-associated lymphoid tissue (GALT), such as Peyer's patches (PPs) and isolated lymphoid follicles, are both histologically and biochemically distinct from normal absorptive epithelial cells of the villi and are termed follicle-associated epithelium (FAE). FAE contains a specialized subset of epithelial cells, M cells, that play an important role in immune surveillance by delivering ingested macromolecules and microorganisms to the underlying lymphoid cells via transcytosis. Despite their significance, studies of M cells remain in their infancy, mainly because the low frequency of M cells and the lack of specific surface markers make it difficult to purify the M cells required for molecular/biochemical analyses. Accordingly, one of the primary aims of our laboratory is to understand the mechanisms that underlie the differentiation and function of FAE and M cells. Our research team is also investigating the recognition of commensal microbiota by intestinal epithelium and its influence on the mucosal and systemic immune system.
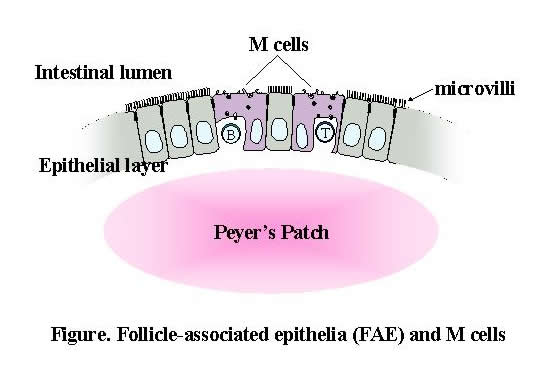
We have devised a method to dissect the epithelial layer from mesenchymes and have thereby collected M-cell-enriched FAE and intestinal epithelial cells (IECs) of the villi from the small intestines of mice. RNA was then extracted from the sampled tissue and amplified. Using Affymetrix high-density oligonucleotide microarrays, we analyzed the gene expression profiles of the FAE and M cells to characterize their cellular phenotypes. The microarray data revealed that among approximately 14,000 genes, 409 were expressed in FAE at more than twice the level observed in the IECs. These included genes that are involved in membrane traffic, host defense, and transcriptional regulation as well as uncharacterized genes. Subsequent real-time RT-PCR and in situ hybridization analyses identified three molecules?ubiquitin D (Ub-D), tumor necrosis factor receptor superfamily 12a (TNFRsf12a), and transmembrane 4 superfamily 4 (Tm4sf4)?that were predominantly distributed throughout FAE but rarely expressed in IECs. In contrast, transcripts of secretory granule neuroendocrine protein 1 (Sgne-1) were scattered in FAE and co-localized with the signal of Ulex europaeus agglutinin-1 (UEA-1); this is relatively specific for M cells in the FAE in mice, although UEA-1 also reacts with goblet cells in villi and Paneth cells in the crypt of the small intestine. This finding clearly suggests the M-cell-specific expression of Sgne-1 in the gut. Such a unique pattern of gene expression distinguishes FAE and M cells from IECs, and may reflect their cellular phenotype(s) associated with specific functional features. We are also currently performing proteome analysis of M cells and FAE, as well as microarray analysis of human biopsy samples obtained by endoscopic examination. This project is being undertaken in collaboration with the RCAI Allergy Transcriptome Research Unit, the Immunogenomics Research Group, the Developmental Genetics Research Group, and Dr. Iimura of Tokyo Women's Medical University.
![]()
The spatial distribution of immune cells in PPs is most likely controlled by chemokine-driven processes. For example, CCL20 is constitutively expressed by the FAE in both mice and humans. Similarly, murine CCL9 and its potential human counterpart CCL23 are selectively produced by FAE but not IECs. The recently identified CXCL16 has dual functions as a transmembrane adhesion molecule and a soluble chemokine. In our research, we found that CXCL16 mRNA and protein were expressed constitutively on the FAE that cover PPs, isolated lymphoid follicles, and cecal patches, but only minimally on IECs within the murine gastrointestinal tract. CXCL16 was also expressed on the FAE of human ileal lymphoid follicles. The CXCL16 receptor CXCR6/Bonzo was constitutively expressed on subpopulations of CD4+ and CD8+ T cells isolated from PPs. The expression of CXCR6/Bonzo on the PP T cells was upregulated after stimulation with anti-CD3 and anti-CD28 mAbs, suggesting that CXCR6 is expressed on the activated/memory PP T cells. The activated PP T cells showed chemotactic migration in response to the soluble N-terminal chemokine domain of CXCL16. Furthermore, the activated PP T cells selectively adhered to those cells expressing CXCL16.To determine the physiological role of CXCL16 in GALT, we first carefully analyzed the distribution of T cells in PPs. We found that T cells are localized in the interfollicular T-cell region (IFR) and to a lesser extent in the subepithelial dome (SED) and the germinal center of lymphoid follicles. Accordingly, the majority of the adoptive transferred activated T cells migrated into the SED and the IFR; however, the neutralization of CXCL16 by anti-CXCL16 antibody injection specifically reduced the migration of the adoptive transferred activated T cells into the SED of PPs. These data suggest that CXCL16 expressed on the FAE plays an important role in the recruitment and retention of activated T cells in the SED, and should therefore be at least partially responsible for lymphocyte compartmentalization in GALT.
In our research, we have identified several M-cell-specific genes in addition to Sgne-1, and we are in the process of determining the precise characteristics of these gene products, including the generation of transgenic and/or knockout mice. We are also attempting to clone the gene for the as-yet unidentified IgA receptor expressed on the M-cell luminal surface. For chemokines expressed by the FAE/M cells, we obtained knockout mice for CXCL16, CCL9, and CCR6 (the receptor for CCL20), and are now analyzing the effect of the absence of these proteins on the differentiation and function of the FAE, M cells, and PPs.We are also interested in host?commensal microbiota interaction. The gut is colonized by some 100 billion commensal bacteria as intestinal microbiota that together with the host cells themselves comprise the host?microbiota symbiotic ecosystem. It is believed that intestinal microbiota is closely related to various lifestyle-related diseases and intestinal infections, and a number intestinal bacteria such as lactobacilli and bifidobacteria are thought to be beneficial against such disease states. Accordingly, consuming these bacteria as probiotics, or functional food, is recommended as a strategy for promoting health and/or preventing disease, although scientific evidence of any benefit in this regard is largely lacking. We are currently addressing this lack of data by using gnotobiotic mice